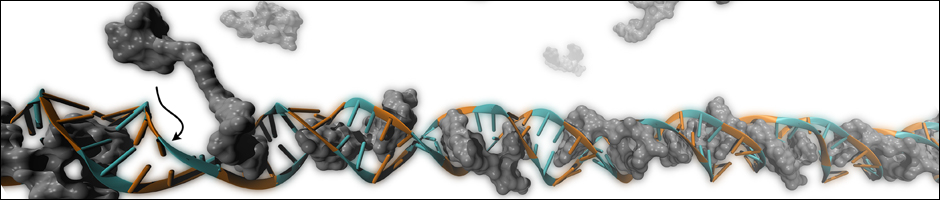
Small Molecule-RNA Binding
Many heritable diseases such as Myotonic Dystrophy, Huntington’s disease, and Fragile X Syndrome are caused by RNA transcripts that contain expanded repeats. In expansions of CUG, CCUG, and CGG, the RNAs are in non-coding regions and the diseases are resulted from RNA gain-of-function. In this mechanism, expanded RNA repeats cause inactivation of proteins, such as MBNL1 and SAM68, which are vital for cell function. In expansion of CAG, expanded repeats are located in coding regions and transcripts are translated as toxic polyQ proteins that cause disease. These RNA repeats have fairly unique elements such as loops with non-Watson-Crick base pairs, which make them ideal targets for pharmacologic development. While researchers have been successful in developing structure-based approaches to drug design for protein targets similar approaches to targeting RNA transcripts are lacking. The dynamic nature of RNA transcripts has made obtaining reasonable predictions for targeting RNA repeats with small molecules a serious challenge.
 Fundamental understanding of these expanded RNA repeats is of upmost importance to the development of drugs that can ameliorate these diseases. Prof. Matthew D. Disney from Scripps Research Institute invented novel approaches for discovering small molecules that bind to RNA repeat expansions and can serve as therapeutics. Yet, the atomistic details of how these small molecules bind to RNA repeats are not clear. In collaboration with the Disney Lab, we utilized different computational methods and predicted how and why such molecules bind specifically to these RNA repeats. The initial results are exciting and promise to advance methods for in silico
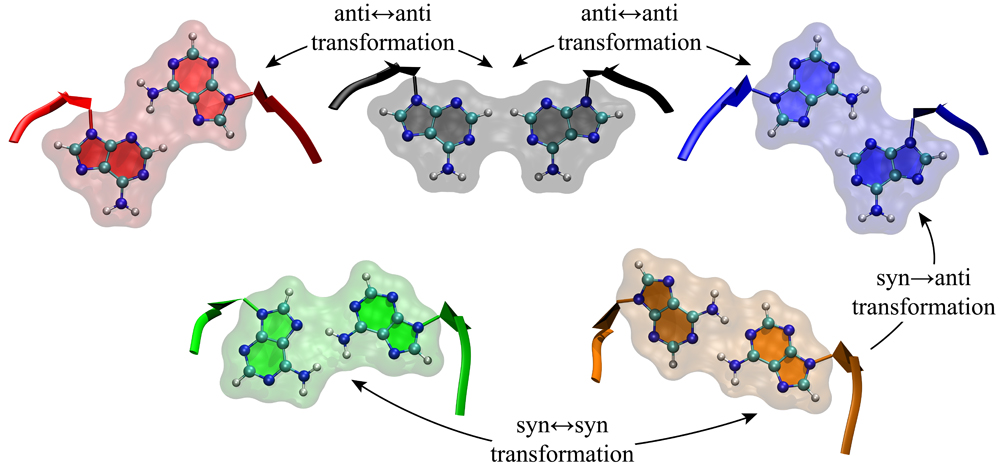
discovery of new therapeutics. We recently showed that 1×1 AA and UU internal loops in RNA CAG and CUG repeat expansions are dynamic and can form
Fundamental understanding of these expanded RNA repeats is of upmost importance to the development of drugs that can ameliorate these diseases. Prof. Matthew D. Disney from Scripps Research Institute invented novel approaches for discovering small molecules that bind to RNA repeat expansions and can serve as therapeutics. Yet, the atomistic details of how these small molecules bind to RNA repeats are not clear. In collaboration with the Disney Lab, we utilized different computational methods and predicted how and why such molecules bind specifically to these RNA repeats. The initial results are exciting and promise to advance methods for in silico
discovery of new therapeutics. We recently showed that 1×1 AA and UU internal loops in RNA CAG and CUG repeat expansions are dynamic and can form 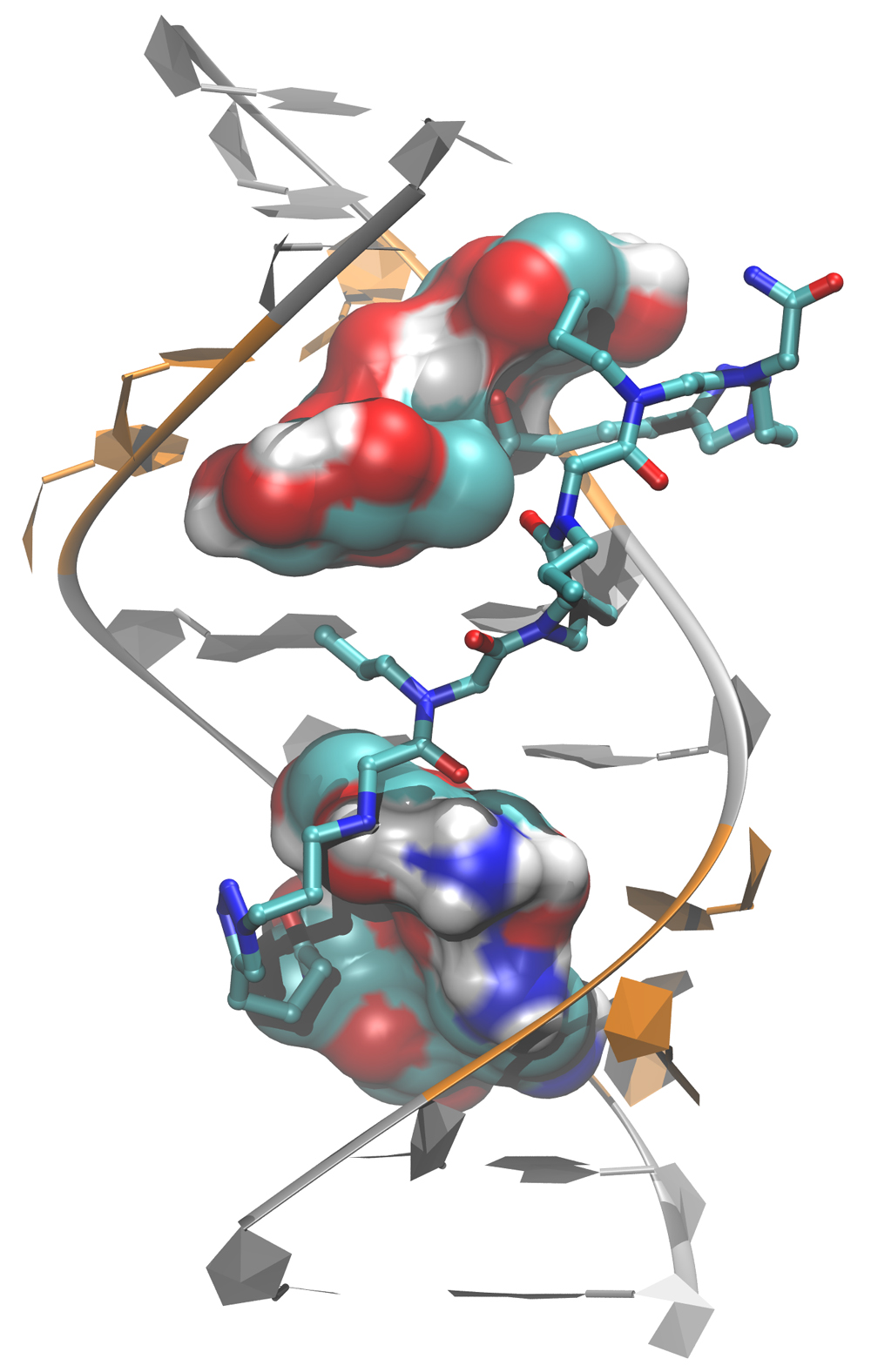 several stable conformations, which could be dominated by AA and UU base pairs upon small molecule binding.
several stable conformations, which could be dominated by AA and UU base pairs upon small molecule binding.
Furthermore, computational studies of RNA CUG and CCUG repeats showed complex transformation pathways in the small molecule binding process. Two press releases were published on these studies discussing the importance of our work in finding cures to Myotonic Dystrophy Type 1 and 2 (NIH Press Release, and Scripps Press Release). Based on these results we hypothesize that the binding of small molecules to RNA repeats causes a change in the global conformation of RNA loops while minimizing the free energies. Collectively, we expect to describe the mechanism behind small molecule RNA binding, and develop next-gen drugs, which can cure RNA-associated diseases. In the long run, the results will have importance in our understanding on other types of roles of RNA in cell such as miRNA and siRNA, which are directly related to cancer. RNA-based therapeutics are getting attention within the pharma industry, such as companies like Isis, Alnylam, and Sarepta trying to develop RNA-based drugs for gene therapy and cancer. Thus, our research will have a big impact in pharmaceutical developments of RNA-based therapeutics.